Multiple data imputation and explainability
RIntroduction
Imputing missing values is quite an important task, but in my experience, very often, it is performed using very simplistic approaches. The basic approach is to impute missing values for numerical features using the average of each feature, or using the mode for categorical features. There are better ways of imputing missing values, for instance by predicting the values using a regression model, or KNN. However, imputing only once is not enough, because each imputed value carries with it a certain level of uncertainty. To account for this, it is better to perform multiple imputation. This means that if you impute your dataset 10 times, you’ll end up with 10 different datasets. Then, you should perform your analysis 10 times, for instance, if training a machine learning model, you should train it on the 10 datasets (and do a train/test split for each, even potentially tune a model for each). Finally, you should pool the results of these 10 analyses.
I have met this approach in the social sciences and statistical literature in general, but very rarely in machine learning. Usually, in the social sciences, explainability is the goal of fitting statistical models to data, and the approach I described above is very well suited for this. Fit 10 (linear) regressions to each imputed dataset, and then pool the estimated coefficients/weights together. Rubin’s rule is used to pool these estimates. You can read more about this rule here. In machine learning, the task is very often prediction; in this case, you should pool the predictions. Computing the average and other statistics of the predictions seem to work just fine in practice.
However, if you are mainly interested in explainability, how should you proceed? I’ve thought a bit about it, and the answer, is “exactly the same way”… I think. What I’m sure about, is you should impute m times, run the analysis m times (which in this case will include getting explanations) and then pool. So the idea is to be able to pool explanations.
Explainability in the “standard” case (no missing values)
To illustrate this idea, I’ll be using the {mice} package for multiple imputation,
{h2o} for the machine learning bit and{iml} for explainability. Note that I could have used
any other machine learning package instead of {h2o} as {iml} is totally package-agnostic.
However, I have been experimenting with {h2o}’s automl implementation lately, so I happened
to have code on hand. Let’s start with the “standard” case where the data does not have any missing
values.
First let’s load the needed packages and initialize h2o functions with h2o.init():
library(tidyverse)
library(Ecdat)
library(mice)
library(h2o)
library(iml)
h2o.init()I’ll be using the DoctorContacts data. Here’s a description:
Click to view the description of the data
DoctorContacts package:Ecdat R Documentation
Contacts With Medical Doctor
Description:
a cross-section from 1977-1978
_number of observations_ : 20186
Usage:
data(DoctorContacts)
Format:
A time serie containing :
mdu number of outpatient visits to a medical doctor
lc log(coinsrate+1) where coinsurance rate is 0 to 100
idp individual deductible plan ?
lpi log(annual participation incentive payment) or 0 if no payment
fmde log(max(medical deductible expenditure)) if IDP=1 and MDE>1
or 0 otherw
physlim physical limitation ?
ndisease number of chronic diseases
health self-rate health (excellent,good,fair,poor)
linc log of annual family income (in \$)
lfam log of family size
educdec years of schooling of household head
age exact age
sex sex (male,female)
child age less than 18 ?
black is household head black ?
Source:
Deb, P. and P.K. Trivedi (2002) “The Structure of Demand for
Medical Care: Latent Class versus Two-Part Models”, _Journal of
Health Economics_, *21*, 601-625.
References:
Cameron, A.C. and P.K. Trivedi (2005) _Microeconometrics :
methods and applications_, Cambridge, pp. 553-556 and 565.The task is to predict "mdu", the number of outpatient visits to an MD. Let’s prepare the data
and split it into 3; a training, validation and holdout set.
data("DoctorContacts")
contacts <- as.h2o(DoctorContacts)splits <- h2o.splitFrame(data=contacts, ratios = c(0.7, 0.2))
original_train <- splits[[1]]
validation <- splits[[2]]
holdout <- splits[[3]]
features_names <- setdiff(colnames(original_train), "mdu")As you see, the ratios argument c(0.7, 0.2) does not add up to 1.
This means that the first of the splits will have 70% of the data, the second split 20% and
the final 10% will be the holdout set.
Let’s first go with a poisson regression. To obtain the same results as with R’s built-in glm()
function, I use the options below, as per H2o’s glm
faq.
If you read Cameron and Trivedi’s Microeconometrics, where this data is presented in the context of count models, you’ll see that they also fit a negative binomial model 2 to this data, as it allows for overdispersion. Here, I’ll stick to a simple poisson regression, simply because the goal of this blog post is not to get the best model; as explained in the beginning, this is an attempt at pooling explanations when doing multiple imputation (and it’s also because GBMs, which I use below, do not handle the negative binomial model).
glm_model <- h2o.glm(y = "mdu", x = features_names,
training_frame = original_train,
validation_frame = validation,
compute_p_values = TRUE,
solver = "IRLSM",
lambda = 0,
remove_collinear_columns = TRUE,
score_each_iteration = TRUE,
family = "poisson",
link = "log")Now that I have this simple model, which returns the (almost) same results as R’s glm() function,
I can take a look at coefficients and see which are important, because GLMs are easily
interpretable:
Click to view
h2o.glm()’s output
summary(glm_model)## Model Details:
## ==============
##
## H2ORegressionModel: glm
## Model Key: GLM_model_R_1572735931328_5
## GLM Model: summary
## family link regularization number_of_predictors_total
## 1 poisson log None 16
## number_of_active_predictors number_of_iterations training_frame
## 1 16 5 RTMP_sid_8588_3
##
## H2ORegressionMetrics: glm
## ** Reported on training data. **
##
## MSE: 17.6446
## RMSE: 4.200547
## MAE: 2.504063
## RMSLE: 0.8359751
## Mean Residual Deviance : 3.88367
## R^2 : 0.1006768
## Null Deviance :64161.44
## Null D.o.F. :14131
## Residual Deviance :54884.02
## Residual D.o.F. :14115
## AIC :83474.52
##
##
## H2ORegressionMetrics: glm
## ** Reported on validation data. **
##
## MSE: 20.85941
## RMSE: 4.56721
## MAE: 2.574582
## RMSLE: 0.8403465
## Mean Residual Deviance : 4.153042
## R^2 : 0.09933874
## Null Deviance :19667.55
## Null D.o.F. :4078
## Residual Deviance :16940.26
## Residual D.o.F. :4062
## AIC :25273.25
##
##
##
##
## Scoring History:
## timestamp duration iterations negative_log_likelihood
## 1 2019-11-03 00:33:46 0.000 sec 0 64161.43611
## 2 2019-11-03 00:33:46 0.004 sec 1 56464.99004
## 3 2019-11-03 00:33:46 0.020 sec 2 54935.05581
## 4 2019-11-03 00:33:47 0.032 sec 3 54884.19756
## 5 2019-11-03 00:33:47 0.047 sec 4 54884.02255
## 6 2019-11-03 00:33:47 0.063 sec 5 54884.02255
## objective
## 1 4.54015
## 2 3.99554
## 3 3.88728
## 4 3.88368
## 5 3.88367
## 6 3.88367
##
## Variable Importances: (Extract with `h2o.varimp`)
## =================================================
##
## variable relative_importance scaled_importance percentage
## 1 black.TRUE 0.67756097 1.00000000 0.236627982
## 2 health.poor 0.48287163 0.71266152 0.168635657
## 3 physlim.TRUE 0.33962316 0.50124369 0.118608283
## 4 health.fair 0.25602066 0.37785627 0.089411366
## 5 sex.male 0.19542639 0.28842628 0.068249730
## 6 ndisease 0.16661902 0.24591001 0.058189190
## 7 idp.TRUE 0.15703578 0.23176627 0.054842384
## 8 child.TRUE 0.09988003 0.14741114 0.034881600
## 9 linc 0.09830075 0.14508030 0.034330059
## 10 lc 0.08126160 0.11993253 0.028379394
## 11 lfam 0.07234463 0.10677213 0.025265273
## 12 fmde 0.06622332 0.09773781 0.023127501
## 13 educdec 0.06416087 0.09469387 0.022407220
## 14 health.good 0.05501613 0.08119732 0.019213558
## 15 age 0.03167598 0.04675000 0.011062359
## 16 lpi 0.01938077 0.02860373 0.006768444As a bonus, let’s see the output of the glm() function:
Click to view
glm()’s output
train_tibble <- as_tibble(original_train)
r_glm <- glm(mdu ~ ., data = train_tibble,
family = poisson(link = "log"))
summary(r_glm)##
## Call:
## glm(formula = mdu ~ ., family = poisson(link = "log"), data = train_tibble)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -5.7039 -1.7890 -0.8433 0.4816 18.4703
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.0005100 0.0585681 0.009 0.9931
## lc -0.0475077 0.0072280 -6.573 4.94e-11 ***
## idpTRUE -0.1794563 0.0139749 -12.841 < 2e-16 ***
## lpi 0.0129742 0.0022141 5.860 4.63e-09 ***
## fmde -0.0166968 0.0042265 -3.951 7.80e-05 ***
## physlimTRUE 0.3182780 0.0126868 25.087 < 2e-16 ***
## ndisease 0.0222300 0.0007215 30.811 < 2e-16 ***
## healthfair 0.2434235 0.0192873 12.621 < 2e-16 ***
## healthgood 0.0231824 0.0115398 2.009 0.0445 *
## healthpoor 0.4608598 0.0329124 14.003 < 2e-16 ***
## linc 0.0826053 0.0062208 13.279 < 2e-16 ***
## lfam -0.1194981 0.0106904 -11.178 < 2e-16 ***
## educdec 0.0205582 0.0019404 10.595 < 2e-16 ***
## age 0.0041397 0.0005152 8.035 9.39e-16 ***
## sexmale -0.2096761 0.0104668 -20.032 < 2e-16 ***
## childTRUE 0.1529588 0.0179179 8.537 < 2e-16 ***
## blackTRUE -0.6231230 0.0176758 -35.253 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for poisson family taken to be 1)
##
## Null deviance: 64043 on 14096 degrees of freedom
## Residual deviance: 55529 on 14080 degrees of freedom
## AIC: 84052
##
## Number of Fisher Scoring iterations: 6I could also use the excellent {ggeffects} package to see the marginal effects of
different variables, for instance "linc":
library(ggeffects)
ggeffect(r_glm, "linc") %>%
ggplot(aes(x, predicted)) +
geom_ribbon(aes(ymin = conf.low, ymax = conf.high), fill = "#0f4150") +
geom_line(colour = "#82518c") +
brotools::theme_blog()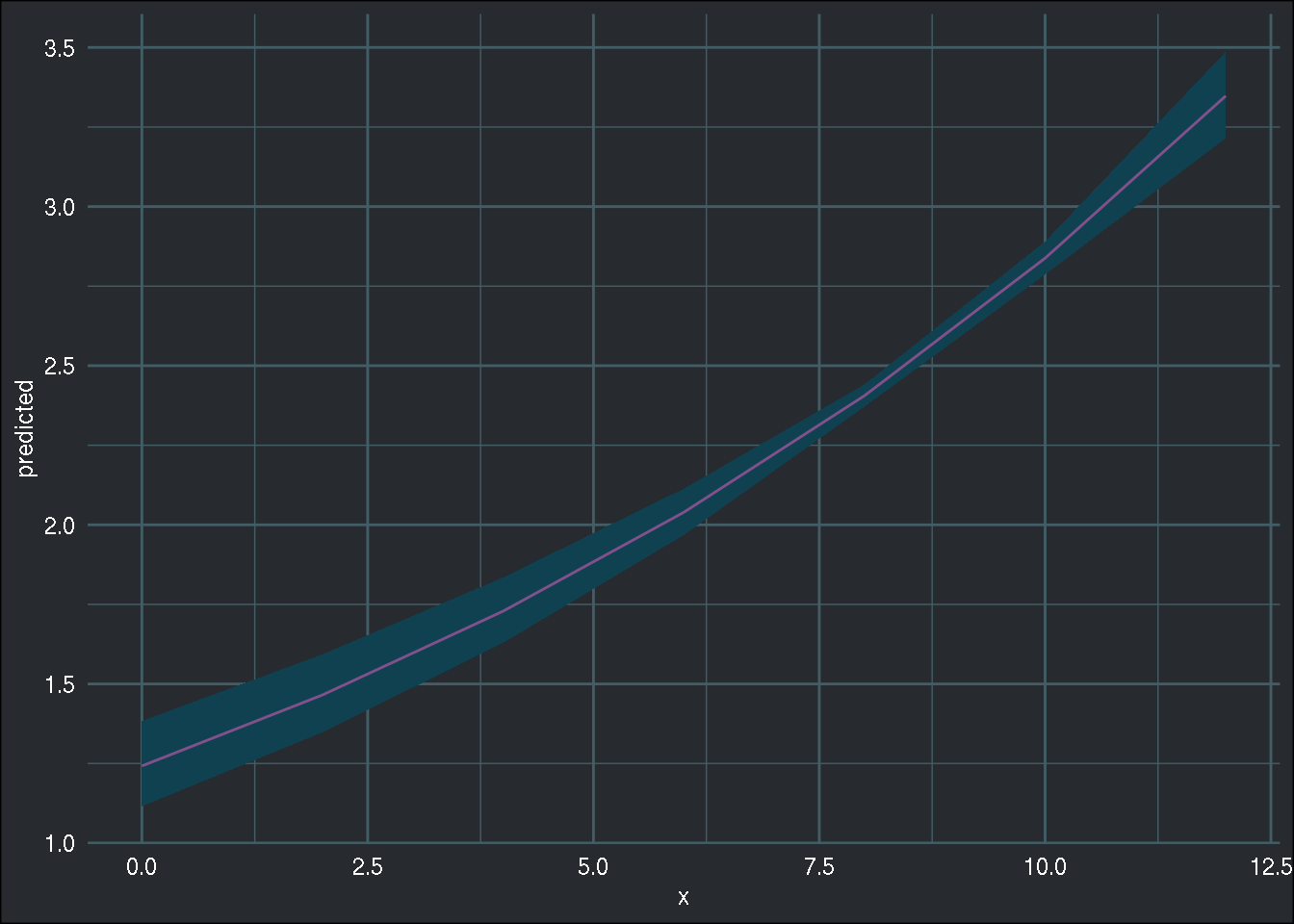
We can see that as “linc” (and other covariates are held constant), the target variable increases.
Let’s also take a look at the marginal effect of a categorical variable, namely "sex":
Click to view another example of marginal effects
library(ggeffects)
ggeffect(r_glm, "sex") %>%
ggplot(aes(x, predicted)) +
geom_point(colour = "#82518c") +
geom_errorbar(aes(x, ymin = conf.low, ymax = conf.high), colour = "#82518c") +
brotools::theme_blog()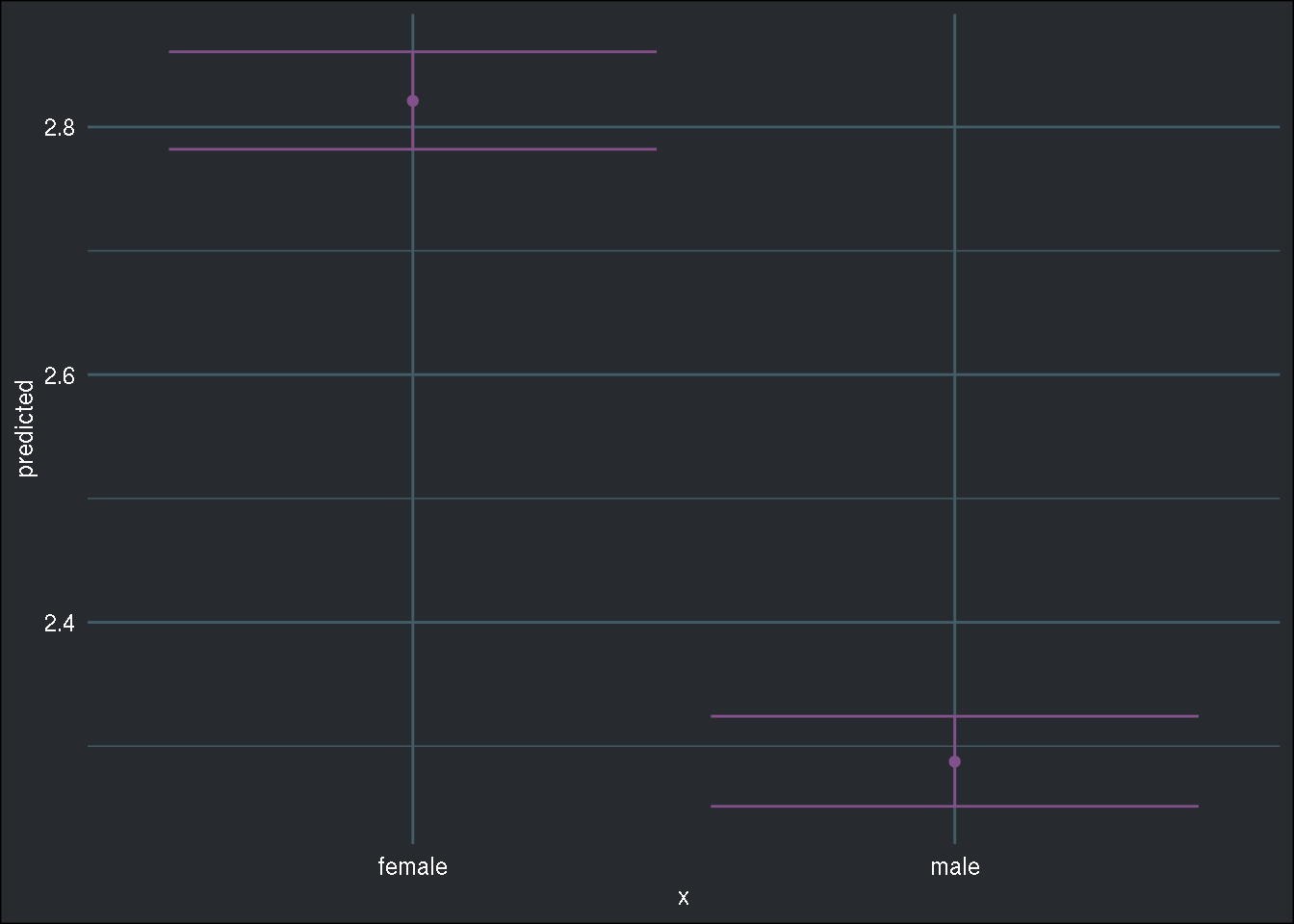
In the case of the "sex" variable, men have significantly less doctor contacts than women.
Now, let’s suppose that I want to train a model with a more complicated name, in order to justify
my salary. Suppose I go with one of those nifty black-box models, for instance a GBM, which
very likely will perform better than the GLM from before. GBMs are available in {h2o} via the
h2o.gbm() function:
gbm_model <- h2o.gbm(y = "mdu", x = features_names,
training_frame = original_train,
validation_frame = validation,
distribution = "poisson",
score_each_iteration = TRUE,
ntrees = 110,
max_depth = 20,
sample_rate = 0.6,
col_sample_rate = 0.8,
col_sample_rate_per_tree = 0.9,
learn_rate = 0.05)To find a set of good hyper-parameter values, I actually used h2o.automl() and then used the
returned parameter values from the leader model. Maybe I’ll write another blog post about
h2o.automl() one day, it’s quite cool. Anyways, now, how do I get me some explainability out of
this? The model does perform better than the GLM as indicated by all the different metrics, but
now I cannot compute any marginal effects, or anything like that. I do get feature importance
by default with:
h2o.varimp(gbm_model)## Variable Importances:
## variable relative_importance scaled_importance percentage
## 1 age 380350.093750 1.000000 0.214908
## 2 linc 282274.343750 0.742143 0.159492
## 3 ndisease 245862.718750 0.646412 0.138919
## 4 lpi 173552.734375 0.456297 0.098062
## 5 educdec 148186.265625 0.389605 0.083729
## 6 lfam 139174.312500 0.365911 0.078637
## 7 fmde 94193.585938 0.247650 0.053222
## 8 health 86160.679688 0.226530 0.048683
## 9 sex 63502.667969 0.166958 0.035881
## 10 lc 50674.968750 0.133232 0.028633
## 11 physlim 45328.382812 0.119175 0.025612
## 12 black 26376.841797 0.069349 0.014904
## 13 idp 24809.185547 0.065227 0.014018
## 14 child 9382.916992 0.024669 0.005302but that’s it. And had I chosen a different “black-box” model, not based on trees, then I would
not even have that.
Thankfully, there’s the amazing {iml} package that contains a lot of functions for model-agnostic
explanations. If you are not familiar with this package and the methods it implements, I highly
encourage you to read the free online ebook
written by the packages author, Christoph Molnar
(who you can follow on Twitter).
Out of the box, {iml} works with several machine learning frameworks, such as {caret} or {mlr}
but not with {h2o}. However, this is not an issue; you only need to create a predict function
which returns a data frame (h2o.predict() used for prediction with h2o models returns an
h2o frame). I have found this interesting blog post from
business-science.io
which explains how to do this. I highly recommend you read this blog post, as it goes much deeper
into the capabilities of {iml}.
So let’s write a predict function that {iml} can use:
#source: https://www.business-science.io/business/2018/08/13/iml-model-interpretability.html
predict_for_iml <- function(model, newdata){
as_tibble(h2o.predict(model, as.h2o(newdata)))
}And let’s now create a Predictor object. These objects are used by {iml} to create explanations:
just_features <- as_tibble(holdout[, 2:15])
actual_target <- as_tibble(holdout[, 1])
predictor_original <- Predictor$new(
model = gbm_model,
data = just_features,
y = actual_target,
predict.fun = predict_for_iml
)predictor_original can now be used to compute all kinds of explanations. I won’t go into much
detail here, as this blog post is already quite long (and I haven’t even reached what I actually
want to write about yet) and you can read more on the before-mentioned blog post or directly
from Christoph Molnar’s ebook linked above.
First, let’s compute a partial dependence plot, which shows the marginal effect of a variable on the outcome. This is to compare it to the one from the GLM model:
feature_effect_original <- FeatureEffect$new(predictor_original, "linc", method = "pdp")
plot(feature_effect_original) +
brotools::theme_blog()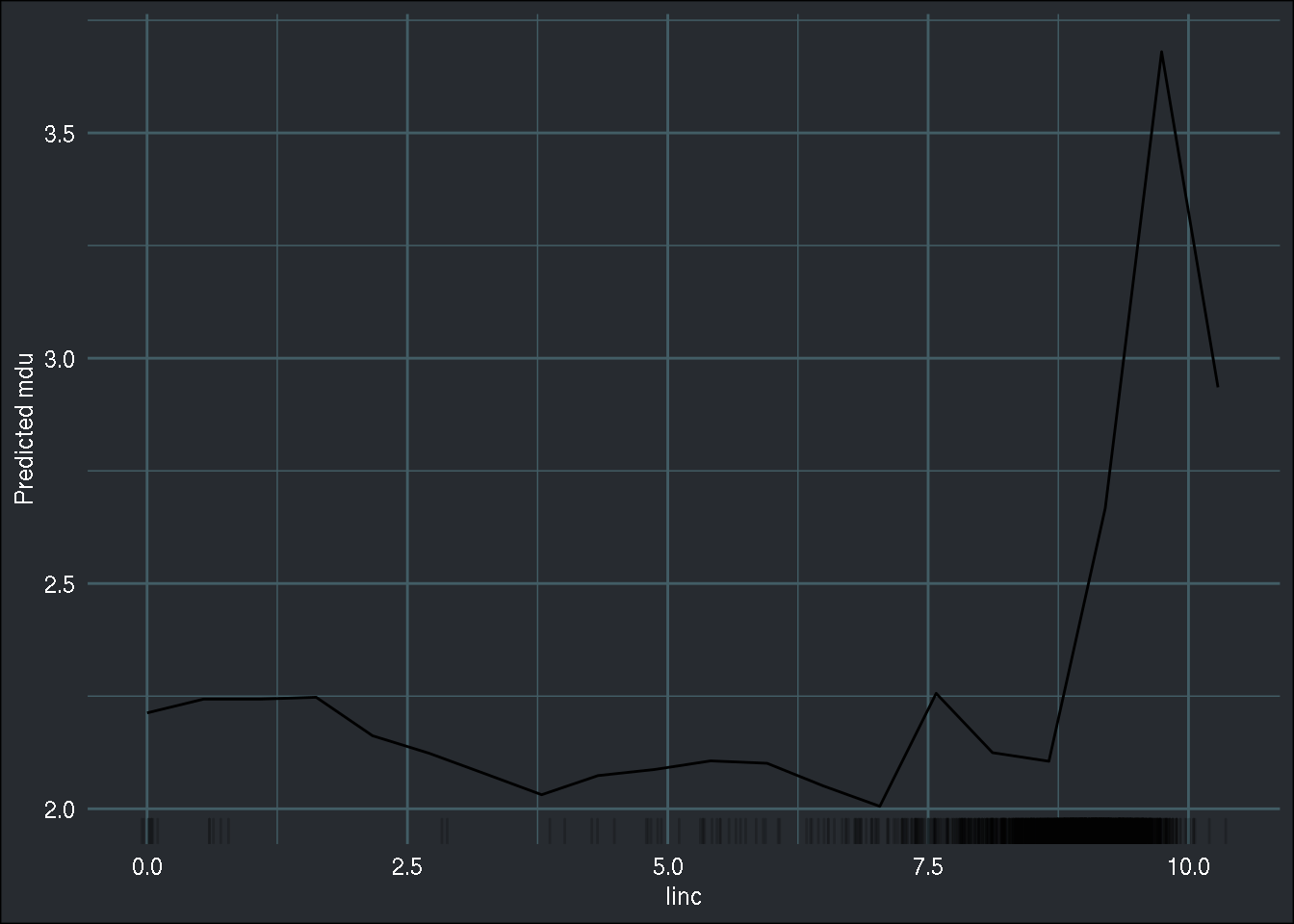
feature_effect_original <- FeatureEffect$new(predictor_original, "linc", method = "pdp")
plot(feature_effect_original) +
brotools::theme_blog()Quite similar to the marginal effects from the GLM! Let’s now compute model-agnostic feature importances:
feature_importance_original <- FeatureImp$new(predictor_original, loss = "mse")
plot(feature_importance_original)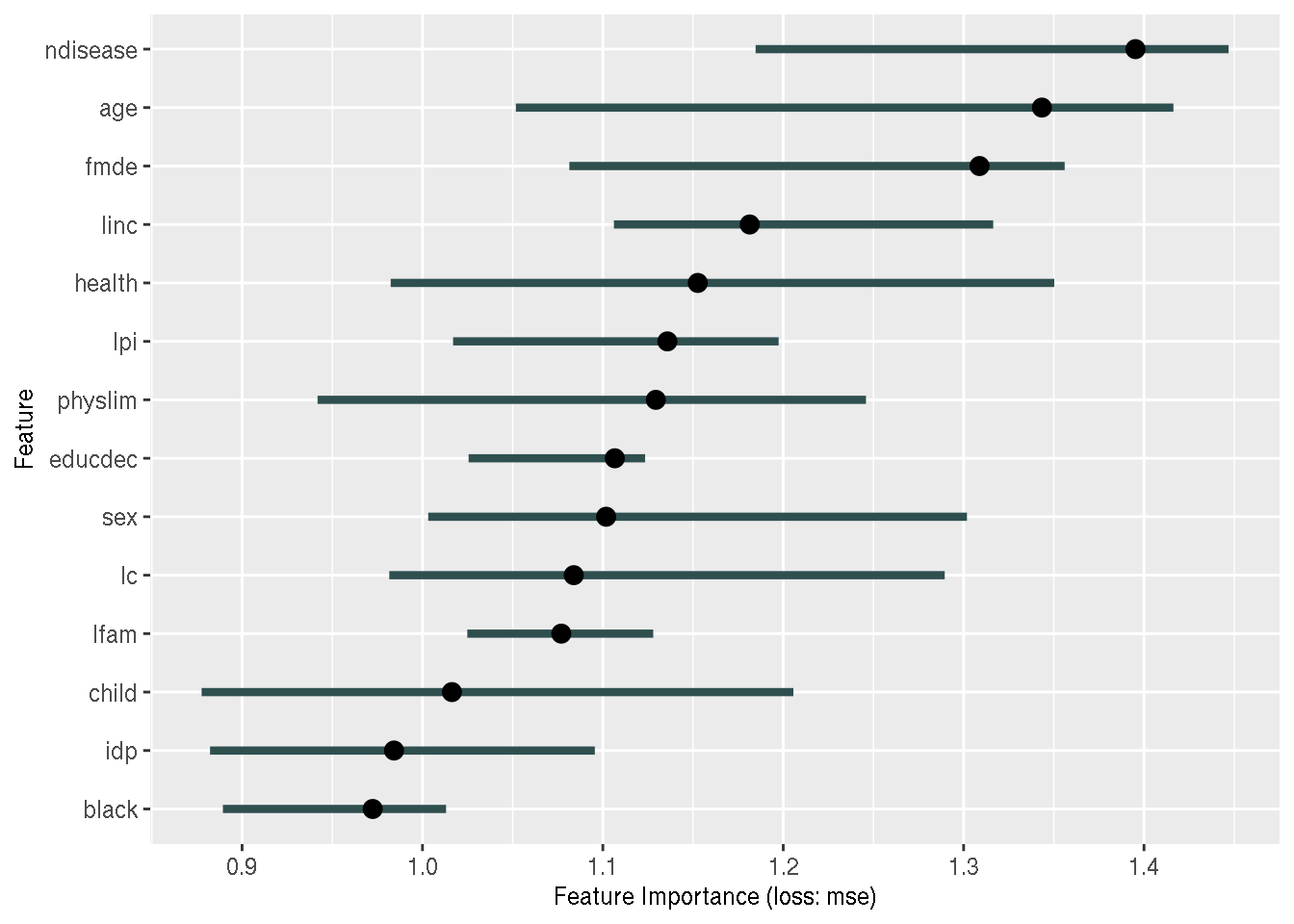
And finally, the interaction effect of the sex variable interacted with all the others:
interaction_sex_original <- Interaction$new(predictor_original, feature = "sex")
plot(interaction_sex_original)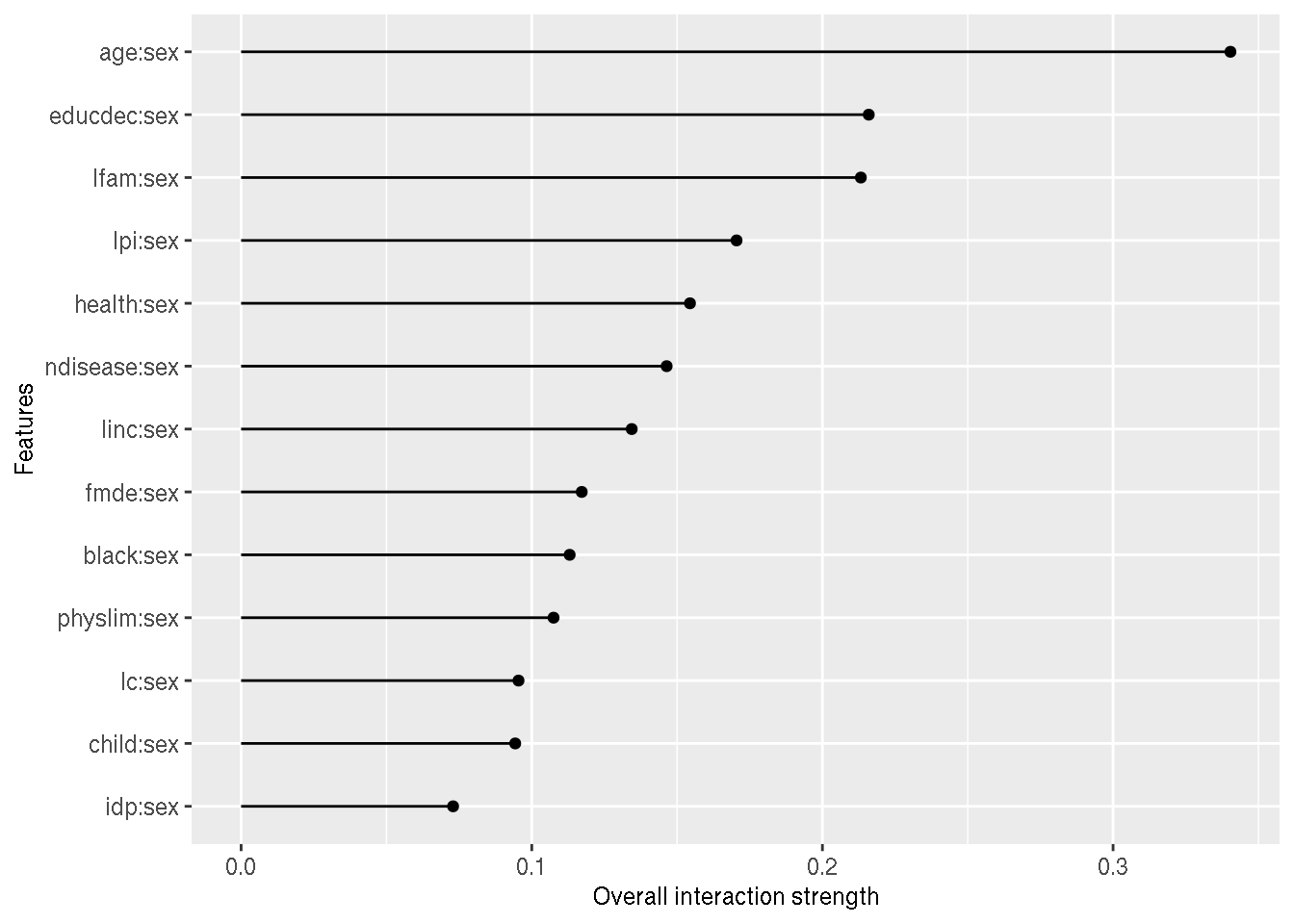
Ok so let’s assume that I’m happy with these explanations, and do need or want to go further. This would be the end of it in an ideal world, but this is not an ideal world unfortunately, but it’s the best we’ve got. In the real world, it often happens that data comes with missing values.
Missing data and explainability
As explained in the beginning, I’ve been wondering how to deal with missing values when the goal of the analysis is explainability. How can the explanations be pooled? Let’s start with creating a data set with missing values, then perform multiple imputation, then perform the analysis.
First, let me create a patterns matrix, that I will pass to the ampute() function from the
{mice} package. This function creates a dataset with missing values, and by using its patterns
argument, I can decide which columns should have missing values:
patterns <- -1*(diag(1, nrow = 15, ncol = 15) - 1)
patterns[ ,c(seq(1, 6), c(9, 13))] <- 0
amputed_train <- ampute(as_tibble(original_train), prop = 0.1, patterns = patterns, mech = "MNAR")## Warning: Data is made numeric because the calculation of weights requires
## numeric dataLet’s take a look at the missingness pattern:
naniar::vis_miss(amputed_train$amp) +
brotools::theme_blog() +
theme(axis.text.x=element_text(angle=90, hjust=1))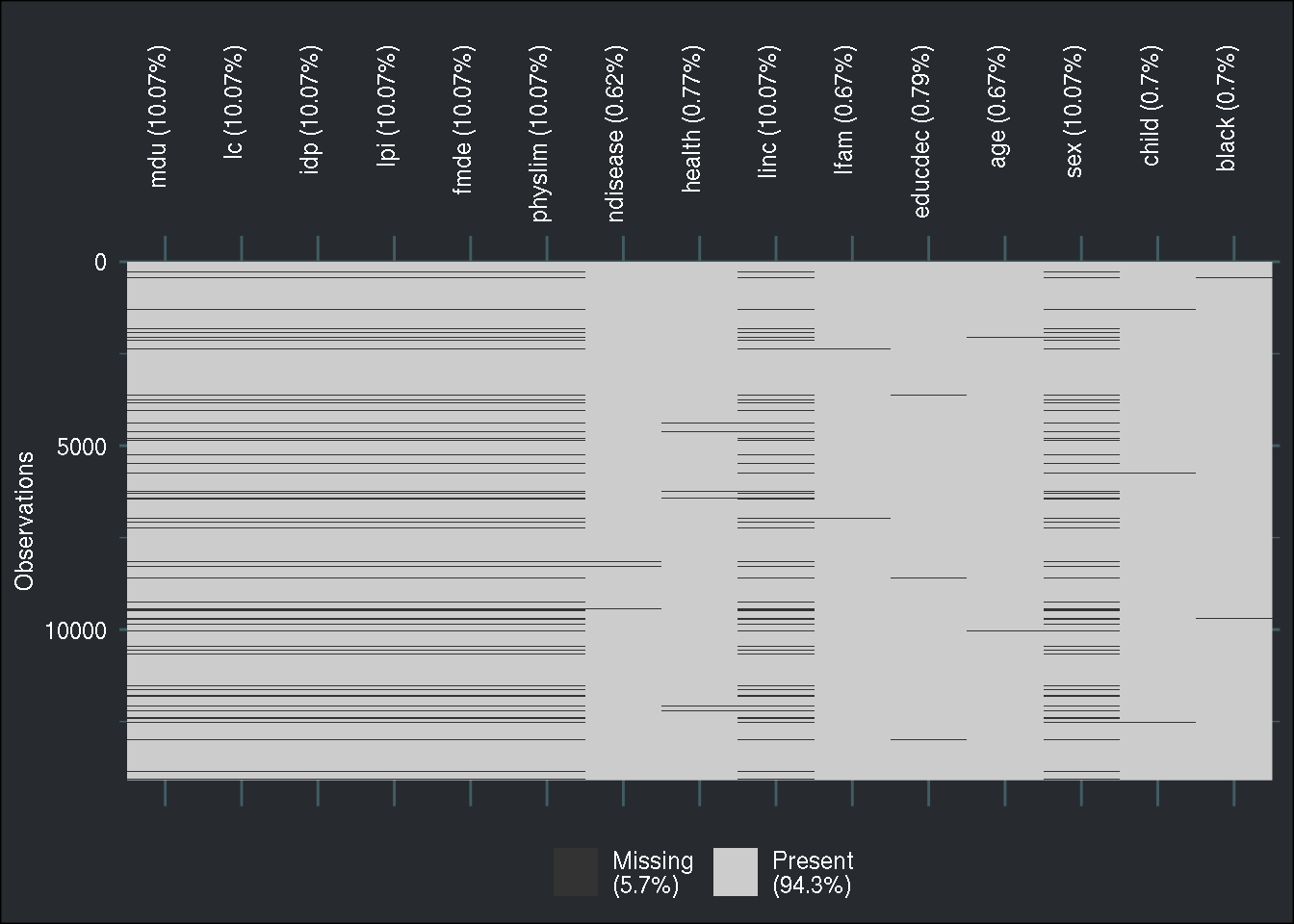
Ok, so now let’s suppose that this was the dataset I was given. As a serious data scientist, I decide to perform multiple imputation first:
imputed_train_data <- mice(data = amputed_train$amp, m = 10)
long_train_data <- complete(imputed_train_data, "long")So because I performed multiple imputation 10 times, I now have 10 different datasets. I should now perform my analysis on these 10 datasets, which means I should run my GBM on each of them, and then get out the explanations for each of them. So let’s do just that. But first, let’s change the columns back to how they were; to perform amputation, the factor columns were converted to numbers:
long_train_data <- long_train_data %>%
mutate(idp = ifelse(idp == 1, FALSE, TRUE),
physlim = ifelse(physlim == 1, FALSE, TRUE),
health = as.factor(case_when(health == 1 ~ "excellent",
health == 2 ~ "fair",
health == 3 ~ "good",
health == 4 ~ "poor")),
sex = as.factor(ifelse(sex == 1, "female", "male")),
child = ifelse(child == 1, FALSE, TRUE),
black = ifelse(black == 1, FALSE, TRUE))Ok, so now we’re ready to go. I will use the h2o.gbm() function on each imputed data set.
For this, I’ll use the group_by()-nest() trick which consists in grouping the dataset by
the .imp column, then nesting it, then mapping the h2o.gbm() function to each imputed
dataset. If you are not familiar with this, you can read
this other blog post, which
explains the approach. I define a custom function, train_on_imputed_data() to run h2o.gbm() on
each imputed data set:
train_on_imputed_data <- function(long_data){
long_data %>%
group_by(.imp) %>%
nest() %>%
mutate(model = map(data, ~h2o.gbm(y = "mdu", x = features_names,
training_frame = as.h2o(.),
validation_frame = validation,
distribution = "poisson",
score_each_iteration = TRUE,
ntrees = 110,
max_depth = 20,
sample_rate = 0.6,
col_sample_rate = 0.8,
col_sample_rate_per_tree = 0.9,
learn_rate = 0.05)))
}Now the training takes place:
imp_trained <- train_on_imputed_data(long_train_data)Let’s take a look at imp_trained:
imp_trained## # A tibble: 10 x 3
## # Groups: .imp [10]
## .imp data model
## <int> <list<df[,16]>> <list>
## 1 1 [14,042 × 16] <H2ORgrsM>
## 2 2 [14,042 × 16] <H2ORgrsM>
## 3 3 [14,042 × 16] <H2ORgrsM>
## 4 4 [14,042 × 16] <H2ORgrsM>
## 5 5 [14,042 × 16] <H2ORgrsM>
## 6 6 [14,042 × 16] <H2ORgrsM>
## 7 7 [14,042 × 16] <H2ORgrsM>
## 8 8 [14,042 × 16] <H2ORgrsM>
## 9 9 [14,042 × 16] <H2ORgrsM>
## 10 10 [14,042 × 16] <H2ORgrsM>We see that the column model contains one model for each imputed dataset. Now comes the
part I wanted to write about (finally): getting explanations out of this. Getting the explanations
from each model is not the hard part, that’s easily done using some {tidyverse} magic (if
you’re following along, run this bit of code below, and go make dinner, have dinner, and
wash the dishes, because it takes time to run):
make_predictors <- function(model){
Predictor$new(
model = model,
data = just_features,
y = actual_target,
predict.fun = predict_for_iml
)
}
make_effect <- function(predictor_object, feature = "linc", method = "pdp"){
FeatureEffect$new(predictor_object, feature, method)
}
make_feat_imp <- function(predictor_object, loss = "mse"){
FeatureImp$new(predictor_object, loss)
}
make_interactions <- function(predictor_object, feature = "sex"){
Interaction$new(predictor_object, feature = feature)
}
imp_trained <- imp_trained %>%
mutate(predictors = map(model, make_predictors)) %>%
mutate(effect_linc = map(predictors, make_effect)) %>%
mutate(feat_imp = map(predictors, make_feat_imp)) %>%
mutate(interactions_sex = map(predictors, make_interactions))Ok so now that I’ve got these explanations, I am done with my analysis. This is the time to pool the results together. Remember, in the case of regression models as used in the social sciences, this means averaging the estimated model parameters and using Rubin’s rule to compute their standard errors. But in this case, this is not so obvious. Should the explanations be averaged? Should I instead analyse them one by one, and see if they differ? My gut feeling is that they shouldn’t differ much, but who knows? Perhaps the answer is doing a bit of both. I have checked online for a paper that would shed some light into this, but have not found any. So let’s take a closer look to the explanations. Let’s look at feature importance:
Click to view the 10 feature importances
imp_trained %>%
pull(feat_imp)## [[1]]
## Interpretation method: FeatureImp
## error function: mse
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## feature importance.05 importance importance.95 permutation.error
## 1 ndisease 1.0421605 1.362672 1.467244 22.03037
## 2 fmde 0.8611917 1.142809 1.258692 18.47583
## 3 lpi 0.8706659 1.103367 1.196081 17.83817
## 4 health 0.8941010 1.098014 1.480508 17.75164
## 5 lc 0.8745229 1.024288 1.296668 16.55970
## 6 black 0.7537278 1.006294 1.095054 16.26879
##
## [[2]]
## Interpretation method: FeatureImp
## error function: mse
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## feature importance.05 importance importance.95 permutation.error
## 1 age 0.984304 1.365702 1.473146 22.52529
## 2 linc 1.102023 1.179169 1.457907 19.44869
## 3 ndisease 1.075821 1.173938 1.642938 19.36241
## 4 fmde 1.059303 1.150112 1.281291 18.96944
## 5 lc 0.837573 1.132719 1.200556 18.68257
## 6 physlim 0.763757 1.117635 1.644434 18.43379
##
## [[3]]
## Interpretation method: FeatureImp
## error function: mse
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## feature importance.05 importance importance.95 permutation.error
## 1 age 0.8641304 1.334382 1.821797 21.62554
## 2 black 1.0553001 1.301338 1.429119 21.09001
## 3 fmde 0.8965085 1.208761 1.360217 19.58967
## 4 ndisease 1.0577766 1.203418 1.651611 19.50309
## 5 linc 0.9299725 1.114041 1.298379 18.05460
## 6 sex 0.9854144 1.091391 1.361406 17.68754
##
## [[4]]
## Interpretation method: FeatureImp
## error function: mse
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## feature importance.05 importance importance.95 permutation.error
## 1 educdec 0.9469049 1.263961 1.358115 20.52909
## 2 age 1.0980269 1.197441 1.763202 19.44868
## 3 health 0.8539843 1.133338 1.343389 18.40753
## 4 linc 0.7608811 1.123423 1.328756 18.24649
## 5 lpi 0.8203850 1.103394 1.251688 17.92118
## 6 black 0.9476909 1.089861 1.328960 17.70139
##
## [[5]]
## Interpretation method: FeatureImp
## error function: mse
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## feature importance.05 importance importance.95 permutation.error
## 1 lpi 0.9897789 1.336405 1.601778 22.03791
## 2 educdec 0.8701162 1.236741 1.424602 20.39440
## 3 age 0.8537786 1.181242 1.261411 19.47920
## 4 lfam 1.0185313 1.133158 1.400151 18.68627
## 5 idp 0.9502284 1.069772 1.203147 17.64101
## 6 linc 0.8600586 1.042453 1.395231 17.19052
##
## [[6]]
## Interpretation method: FeatureImp
## error function: mse
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## feature importance.05 importance importance.95 permutation.error
## 1 lc 0.7707383 1.208190 1.379422 19.65436
## 2 sex 0.9309901 1.202629 1.479511 19.56391
## 3 linc 1.0549563 1.138404 1.624217 18.51912
## 4 lpi 0.9360817 1.135198 1.302084 18.46696
## 5 physlim 0.7357272 1.132525 1.312584 18.42349
## 6 child 1.0199964 1.109120 1.316306 18.04274
##
## [[7]]
## Interpretation method: FeatureImp
## error function: mse
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## feature importance.05 importance importance.95 permutation.error
## 1 linc 0.9403425 1.262994 1.511122 20.65942
## 2 lc 1.0481333 1.233136 1.602796 20.17103
## 3 ndisease 1.1612194 1.212454 1.320208 19.83272
## 4 educdec 0.7924637 1.197343 1.388218 19.58554
## 5 lfam 0.8423790 1.178545 1.349884 19.27805
## 6 age 0.9125829 1.168297 1.409525 19.11043
##
## [[8]]
## Interpretation method: FeatureImp
## error function: mse
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## feature importance.05 importance importance.95 permutation.error
## 1 age 1.1281736 1.261273 1.609524 20.55410
## 2 health 0.9134557 1.240597 1.432366 20.21716
## 3 lfam 0.7469043 1.182294 1.345910 19.26704
## 4 lpi 0.8088552 1.160863 1.491139 18.91779
## 5 ndisease 1.0756671 1.104357 1.517278 17.99695
## 6 fmde 0.6929092 1.093465 1.333544 17.81946
##
## [[9]]
## Interpretation method: FeatureImp
## error function: mse
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## feature importance.05 importance importance.95 permutation.error
## 1 educdec 1.0188109 1.287697 1.381982 20.92713
## 2 lpi 0.9853336 1.213095 1.479002 19.71473
## 3 linc 0.8354715 1.195344 1.254350 19.42625
## 4 age 0.9980451 1.179371 1.383545 19.16666
## 5 ndisease 1.0492685 1.176804 1.397398 19.12495
## 6 lfam 1.0814043 1.166626 1.264592 18.95953
##
## [[10]]
## Interpretation method: FeatureImp
## error function: mse
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## feature importance.05 importance importance.95 permutation.error
## 1 age 0.9538824 1.211869 1.621151 19.53671
## 2 sex 0.9148921 1.211253 1.298311 19.52678
## 3 lfam 0.8227355 1.093094 1.393815 17.62192
## 4 ndisease 0.8282127 1.090779 1.205994 17.58459
## 5 lc 0.7004401 1.060870 1.541697 17.10244
## 6 health 0.8137149 1.058324 1.183639 17.06138As you can see, the feature importances are quite different from each other, but I don’t think this comes from the imputations, but rather from the fact that feature importance depends on shuffling the feature, which adds randomness to the measurement (source: https://christophm.github.io/interpretable-ml-book/feature-importance.html#disadvantages-9). To mitigate this, Christoph Molnar suggests repeating the the permutation and averaging the importance measures; I think that this would be my approach for pooling as well.
Let’s now take a look at interactions:
Click to view the 10 interactions
imp_trained %>%
pull(interactions_sex)## [[1]]
## Interpretation method: Interaction
##
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## .feature .interaction
## 1 lc:sex 0.07635197
## 2 idp:sex 0.08172754
## 3 lpi:sex 0.10704357
## 4 fmde:sex 0.11267146
## 5 physlim:sex 0.04099073
## 6 ndisease:sex 0.16314524
##
## [[2]]
## Interpretation method: Interaction
##
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## .feature .interaction
## 1 lc:sex 0.10349820
## 2 idp:sex 0.07432519
## 3 lpi:sex 0.11651413
## 4 fmde:sex 0.18123926
## 5 physlim:sex 0.12952808
## 6 ndisease:sex 0.14528876
##
## [[3]]
## Interpretation method: Interaction
##
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## .feature .interaction
## 1 lc:sex 0.05919320
## 2 idp:sex 0.05586197
## 3 lpi:sex 0.24253335
## 4 fmde:sex 0.05240474
## 5 physlim:sex 0.06404969
## 6 ndisease:sex 0.14508072
##
## [[4]]
## Interpretation method: Interaction
##
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## .feature .interaction
## 1 lc:sex 0.02775529
## 2 idp:sex 0.02050390
## 3 lpi:sex 0.11781130
## 4 fmde:sex 0.11084240
## 5 physlim:sex 0.17932694
## 6 ndisease:sex 0.07181589
##
## [[5]]
## Interpretation method: Interaction
##
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## .feature .interaction
## 1 lc:sex 0.12873151
## 2 idp:sex 0.03681428
## 3 lpi:sex 0.15879389
## 4 fmde:sex 0.16952900
## 5 physlim:sex 0.07031520
## 6 ndisease:sex 0.10567463
##
## [[6]]
## Interpretation method: Interaction
##
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## .feature .interaction
## 1 lc:sex 0.15320481
## 2 idp:sex 0.08645037
## 3 lpi:sex 0.16674641
## 4 fmde:sex 0.14671054
## 5 physlim:sex 0.09236257
## 6 ndisease:sex 0.14605618
##
## [[7]]
## Interpretation method: Interaction
##
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## .feature .interaction
## 1 lc:sex 0.04072960
## 2 idp:sex 0.05641868
## 3 lpi:sex 0.19491959
## 4 fmde:sex 0.07119644
## 5 physlim:sex 0.05777469
## 6 ndisease:sex 0.16555363
##
## [[8]]
## Interpretation method: Interaction
##
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## .feature .interaction
## 1 lc:sex 0.04979709
## 2 idp:sex 0.06036898
## 3 lpi:sex 0.14009307
## 4 fmde:sex 0.10927688
## 5 physlim:sex 0.08761533
## 6 ndisease:sex 0.20544585
##
## [[9]]
## Interpretation method: Interaction
##
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## .feature .interaction
## 1 lc:sex 0.08572075
## 2 idp:sex 0.12254979
## 3 lpi:sex 0.17532347
## 4 fmde:sex 0.12557420
## 5 physlim:sex 0.05084209
## 6 ndisease:sex 0.13977328
##
## [[10]]
## Interpretation method: Interaction
##
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## .feature .interaction
## 1 lc:sex 0.08636490
## 2 idp:sex 0.04807331
## 3 lpi:sex 0.17922280
## 4 fmde:sex 0.05728403
## 5 physlim:sex 0.09392774
## 6 ndisease:sex 0.13408956It would seem that interactions are a bit more stable. Let’s average the values; for this
I need to access the results element of the interactions object and the result out:
interactions_sex_result <- imp_trained %>%
mutate(interactions_results = map(interactions_sex, function(x)(x$results))) %>%
pull()interactions_sex_result is a list of dataframes, which means I can bind the rows together and
compute whatever I need:
interactions_sex_result %>%
bind_rows() %>%
group_by(.feature) %>%
summarise_at(.vars = vars(.interaction),
.funs = funs(mean, sd, low_ci = quantile(., 0.05), high_ci = quantile(., 0.95)))## # A tibble: 13 x 5
## .feature mean sd low_ci high_ci
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 age:sex 0.294 0.0668 0.181 0.369
## 2 black:sex 0.117 0.0286 0.0763 0.148
## 3 child:sex 0.0817 0.0308 0.0408 0.125
## 4 educdec:sex 0.148 0.0411 0.104 0.220
## 5 fmde:sex 0.114 0.0443 0.0546 0.176
## 6 health:sex 0.130 0.0190 0.104 0.151
## 7 idp:sex 0.0643 0.0286 0.0278 0.106
## 8 lc:sex 0.0811 0.0394 0.0336 0.142
## 9 lfam:sex 0.149 0.0278 0.125 0.198
## 10 linc:sex 0.142 0.0277 0.104 0.179
## 11 lpi:sex 0.160 0.0416 0.111 0.221
## 12 ndisease:sex 0.142 0.0356 0.0871 0.187
## 13 physlim:sex 0.0867 0.0415 0.0454 0.157That seems pretty good. Now, what about the partial dependence? Let’s take a closer look:
Click to view the 10 pdps
imp_trained %>%
pull(effect_linc)## [[1]]
## Interpretation method: FeatureEffect
## features: linc[numerical]
## grid size: 20
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## linc .y.hat .type
## 1 0.0000000 1.652445 pdp
## 2 0.5312226 1.687522 pdp
## 3 1.0624453 1.687522 pdp
## 4 1.5936679 1.687522 pdp
## 5 2.1248905 1.685088 pdp
## 6 2.6561132 1.694112 pdp
##
## [[2]]
## Interpretation method: FeatureEffect
## features: linc[numerical]
## grid size: 20
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## linc .y.hat .type
## 1 0.0000000 1.813449 pdp
## 2 0.5312226 1.816195 pdp
## 3 1.0624453 1.816195 pdp
## 4 1.5936679 1.816195 pdp
## 5 2.1248905 1.804457 pdp
## 6 2.6561132 1.797238 pdp
##
## [[3]]
## Interpretation method: FeatureEffect
## features: linc[numerical]
## grid size: 20
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## linc .y.hat .type
## 1 0.0000000 1.906515 pdp
## 2 0.5312226 2.039318 pdp
## 3 1.0624453 2.039318 pdp
## 4 1.5936679 2.039318 pdp
## 5 2.1248905 2.002970 pdp
## 6 2.6561132 2.000922 pdp
##
## [[4]]
## Interpretation method: FeatureEffect
## features: linc[numerical]
## grid size: 20
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## linc .y.hat .type
## 1 0.0000000 1.799552 pdp
## 2 0.5312226 2.012634 pdp
## 3 1.0624453 2.012634 pdp
## 4 1.5936679 2.012634 pdp
## 5 2.1248905 1.982425 pdp
## 6 2.6561132 1.966392 pdp
##
## [[5]]
## Interpretation method: FeatureEffect
## features: linc[numerical]
## grid size: 20
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## linc .y.hat .type
## 1 0.0000000 1.929158 pdp
## 2 0.5312226 1.905171 pdp
## 3 1.0624453 1.905171 pdp
## 4 1.5936679 1.905171 pdp
## 5 2.1248905 1.879721 pdp
## 6 2.6561132 1.869113 pdp
##
## [[6]]
## Interpretation method: FeatureEffect
## features: linc[numerical]
## grid size: 20
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## linc .y.hat .type
## 1 0.0000000 2.147697 pdp
## 2 0.5312226 2.162393 pdp
## 3 1.0624453 2.162393 pdp
## 4 1.5936679 2.162393 pdp
## 5 2.1248905 2.119923 pdp
## 6 2.6561132 2.115131 pdp
##
## [[7]]
## Interpretation method: FeatureEffect
## features: linc[numerical]
## grid size: 20
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## linc .y.hat .type
## 1 0.0000000 1.776742 pdp
## 2 0.5312226 1.957938 pdp
## 3 1.0624453 1.957938 pdp
## 4 1.5936679 1.957938 pdp
## 5 2.1248905 1.933847 pdp
## 6 2.6561132 1.885287 pdp
##
## [[8]]
## Interpretation method: FeatureEffect
## features: linc[numerical]
## grid size: 20
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## linc .y.hat .type
## 1 0.0000000 2.020647 pdp
## 2 0.5312226 2.017981 pdp
## 3 1.0624453 2.017981 pdp
## 4 1.5936679 2.017981 pdp
## 5 2.1248905 1.981122 pdp
## 6 2.6561132 2.017604 pdp
##
## [[9]]
## Interpretation method: FeatureEffect
## features: linc[numerical]
## grid size: 20
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## linc .y.hat .type
## 1 0.0000000 1.811189 pdp
## 2 0.5312226 2.003053 pdp
## 3 1.0624453 2.003053 pdp
## 4 1.5936679 2.003053 pdp
## 5 2.1248905 1.938150 pdp
## 6 2.6561132 1.918518 pdp
##
## [[10]]
## Interpretation method: FeatureEffect
## features: linc[numerical]
## grid size: 20
##
## Analysed predictor:
## Prediction task: unknown
##
##
## Analysed data:
## Sampling from data.frame with 2013 rows and 14 columns.
##
## Head of results:
## linc .y.hat .type
## 1 0.0000000 1.780325 pdp
## 2 0.5312226 1.850203 pdp
## 3 1.0624453 1.850203 pdp
## 4 1.5936679 1.850203 pdp
## 5 2.1248905 1.880805 pdp
## 6 2.6561132 1.881305 pdpAs you can see, the values are quite similar. I think that in the case of plots, the best way to visualize the impact of the imputation is to simply plot all the lines in a single plot:
effect_linc_results <- imp_trained %>%
mutate(effect_linc_results = map(effect_linc, function(x)(x$results))) %>%
select(.imp, effect_linc_results) %>%
unnest(effect_linc_results)
effect_linc_results %>%
bind_rows() %>%
ggplot() +
geom_line(aes(y = .y.hat, x = linc, group = .imp), colour = "#82518c") +
brotools::theme_blog()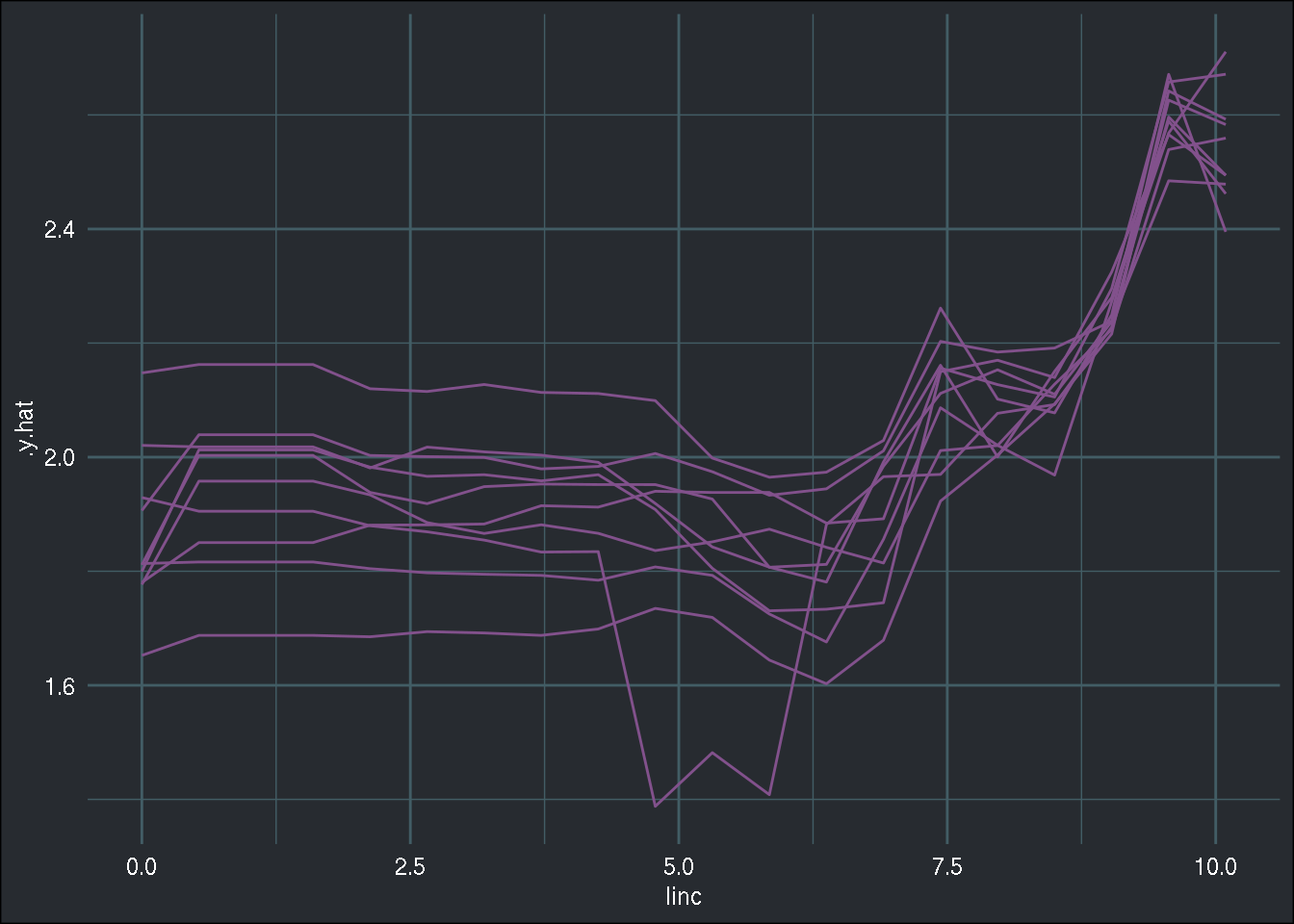
Overall, the partial dependence plot seems to behave in a very similar way across the different imputed datasets!
To conclude, I think that the approach I suggest here is nothing revolutionary; it is consistent with the way one should conduct an analysis with multiple imputed datasets. However, the pooling step is non-trivial and there is no magic recipe; it really depends on the goal of the analysis and what you want or need to show.
Hope you enjoyed! If you found this blog post useful, you might want to follow me on twitter for blog post updates and buy me an espresso or paypal.me, or buy my ebook on Leanpub.

